Help/FAQ
Quick Search
In CovPDB, you can quickly view the list of all ligands, proteins, complexes, warheads, covalent mechanisms, and targetable residues by clicking one of the six main representative icons on the homepage.
Browse CovPDB
Ligands
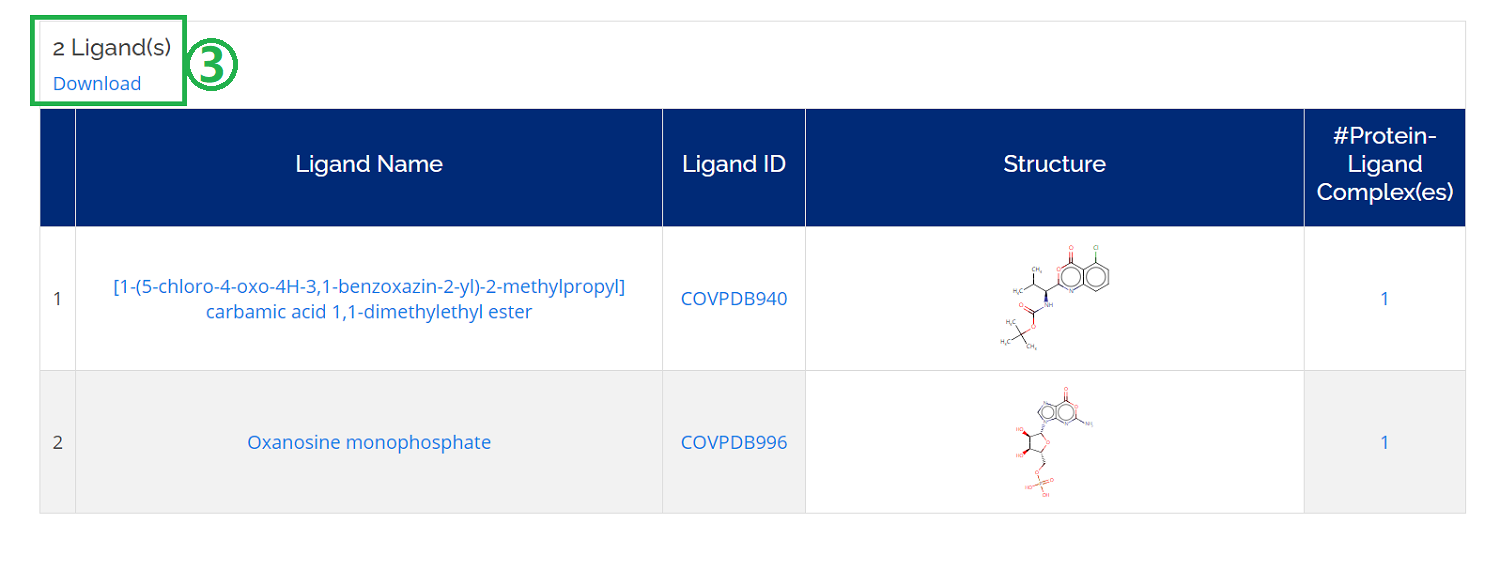
There is the possibility to search for compounds ordered alphabetically. To do so, navigate to Home -> Browse -> Ligands (List) and you could also find the related information of the ligand, like structures and the number of complexes.
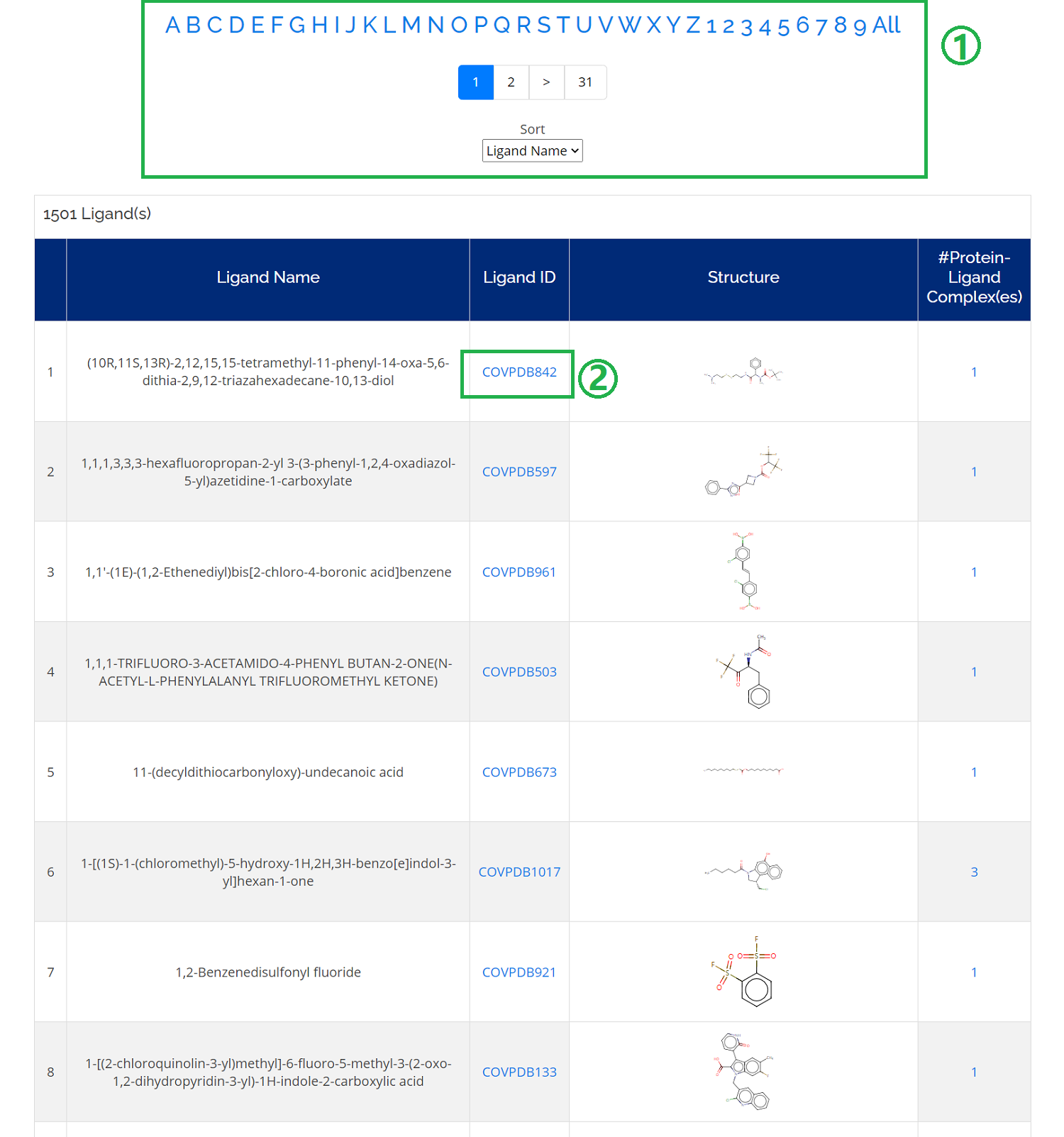
- 1. You can sort the ligand list alphabetically or sort list according to Ligand Name, Ligand ID, and #Complexes;
- 2. Check detailed information by clicking the ligand ID .
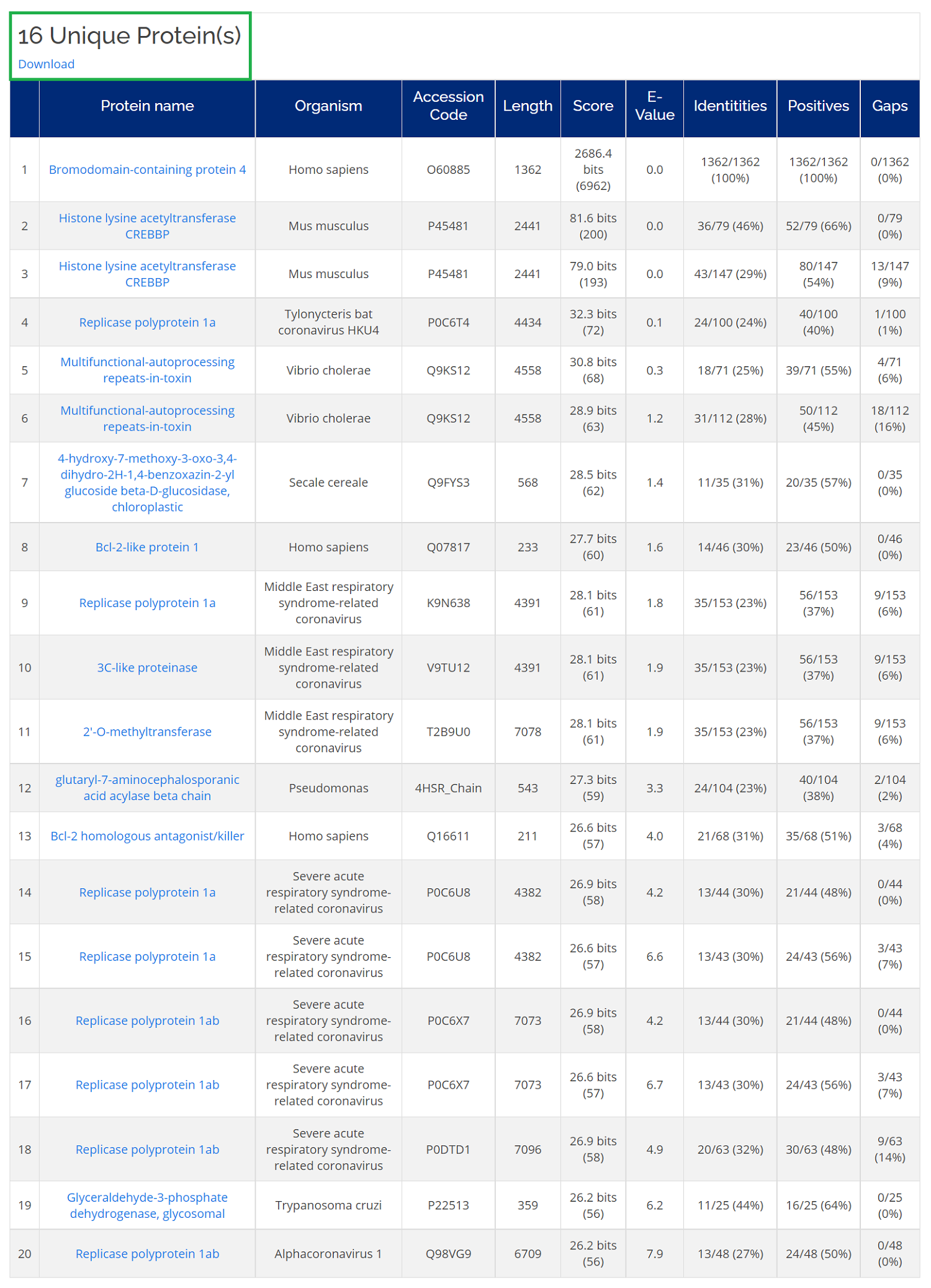
Proteins
Click on the Proteins icon on the homepage or Home -> Browse -> Proteins.
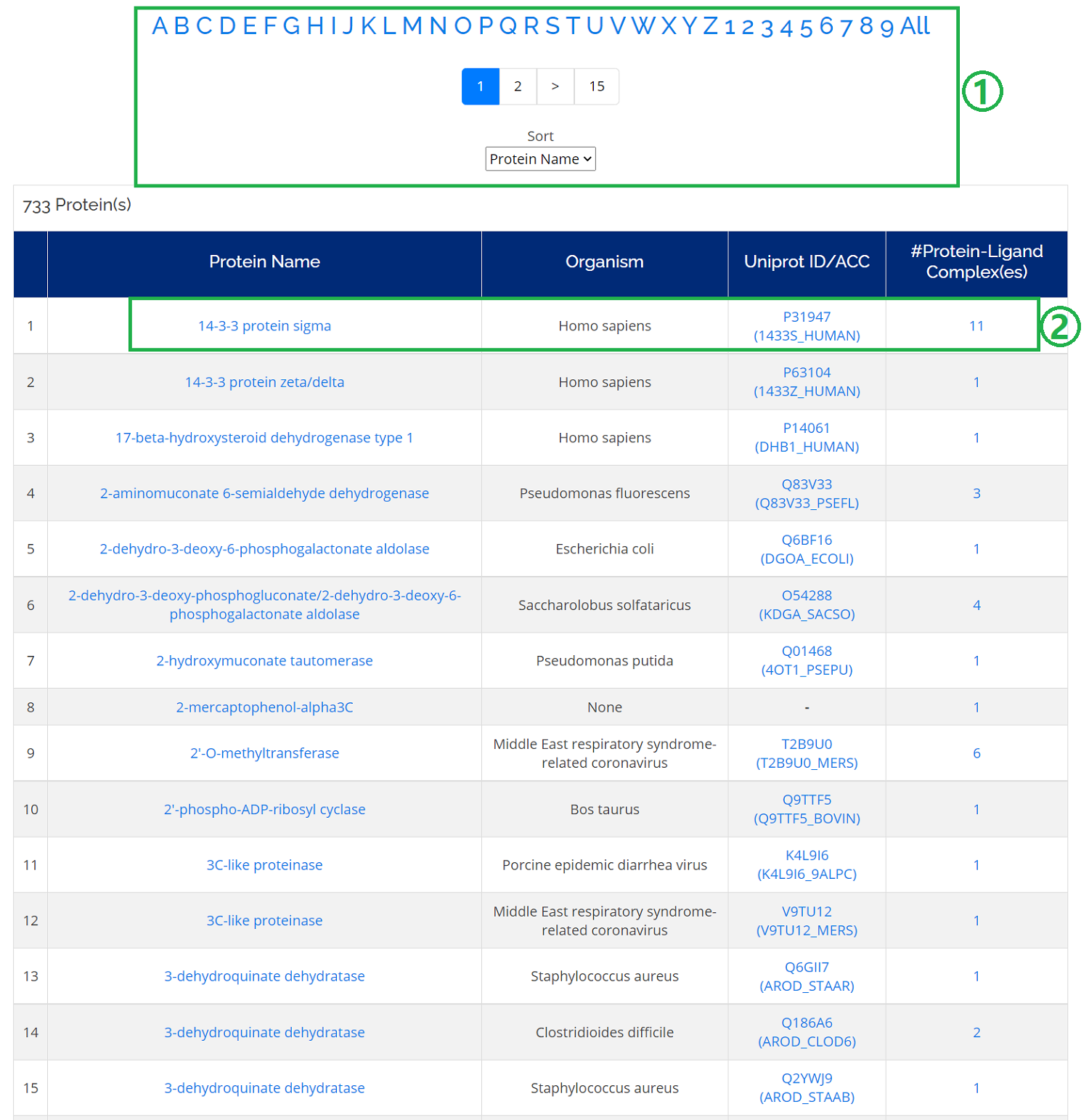
- 1. You can sort the protein list according to Protein Name, Uniprot ID, Organism, and #Complexes;
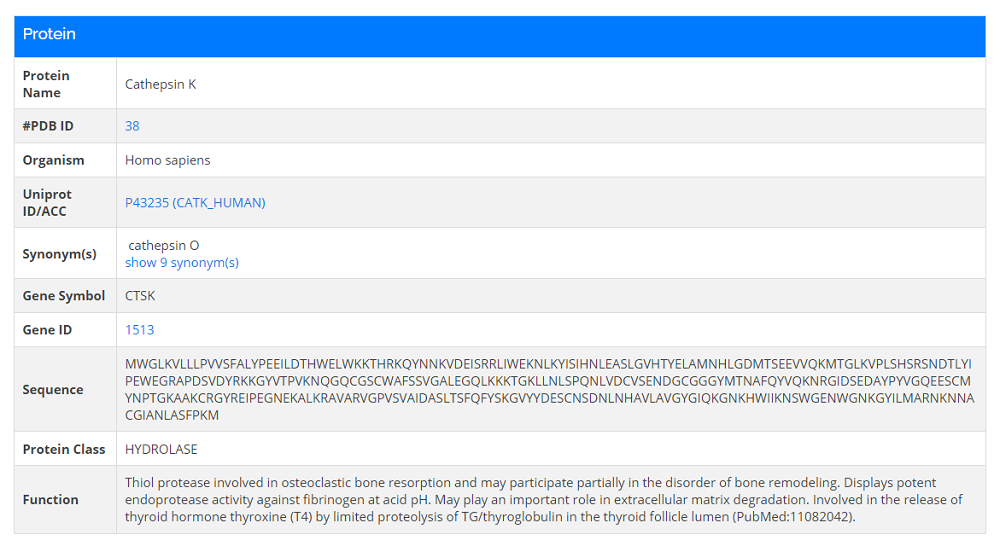
- 2. Click any protein name in the protein list and a protein card will be opened.
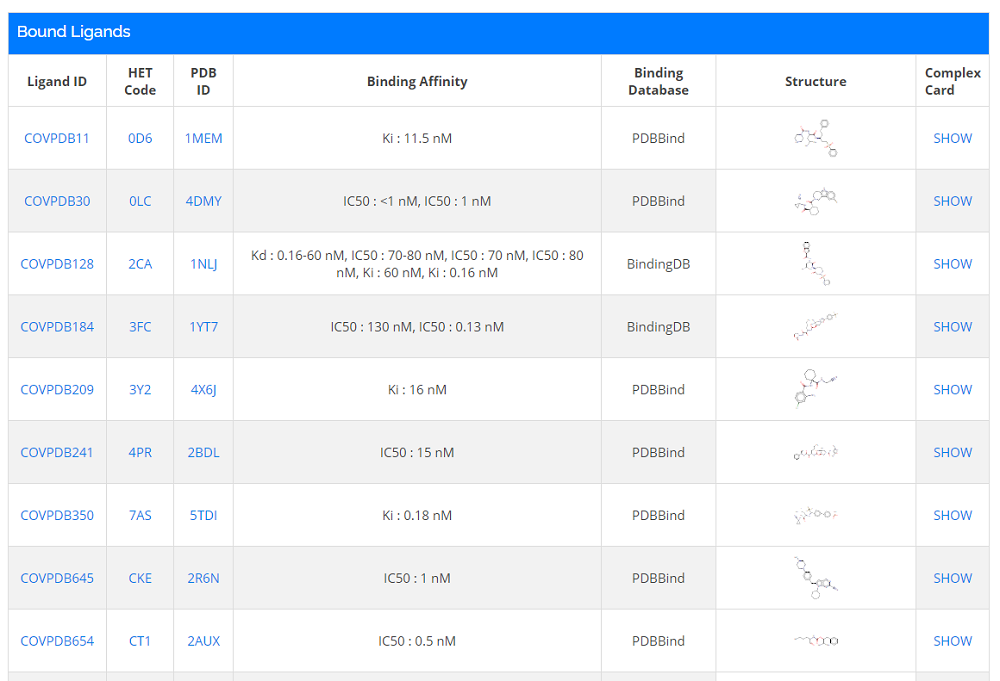
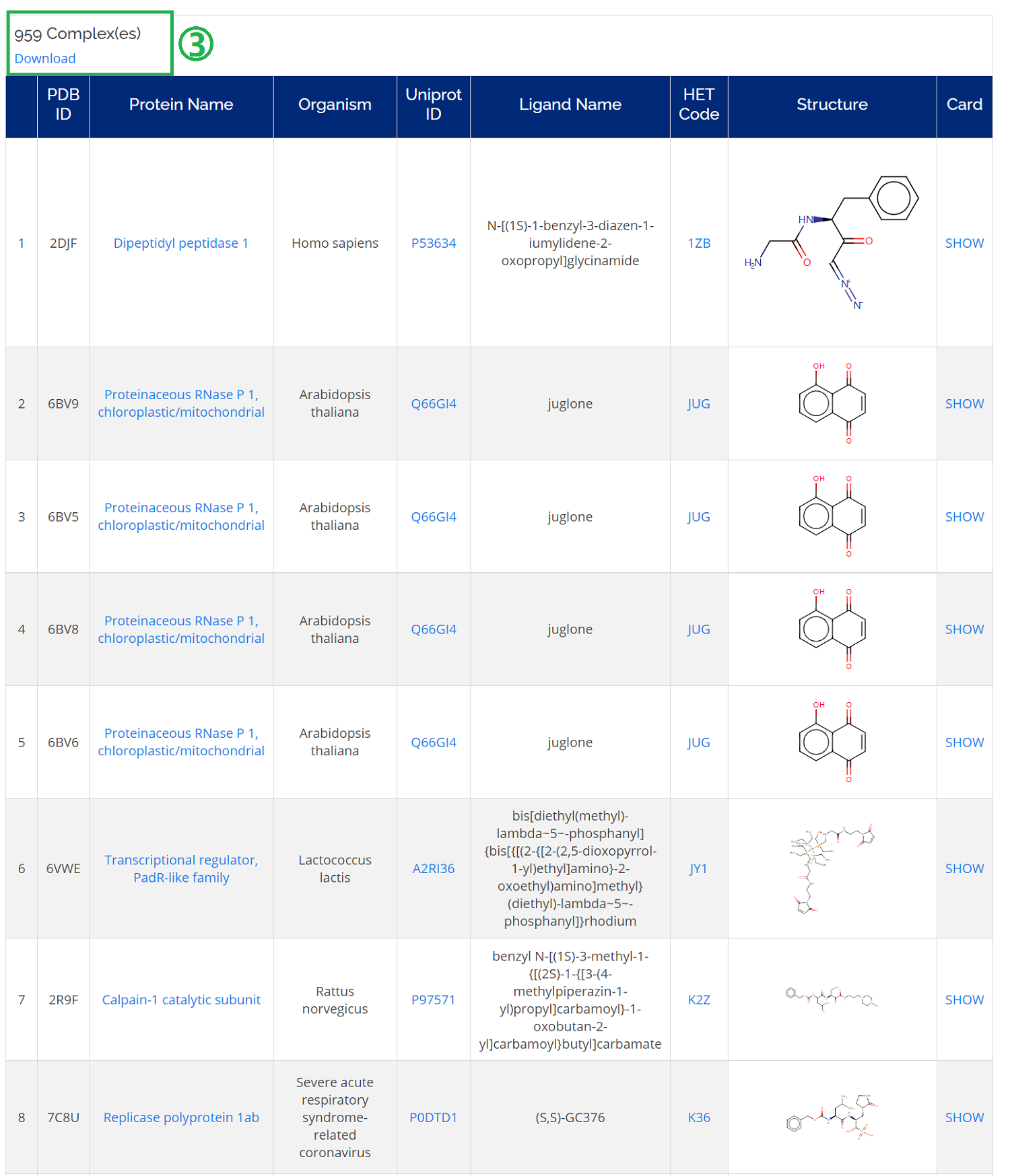
Complexes
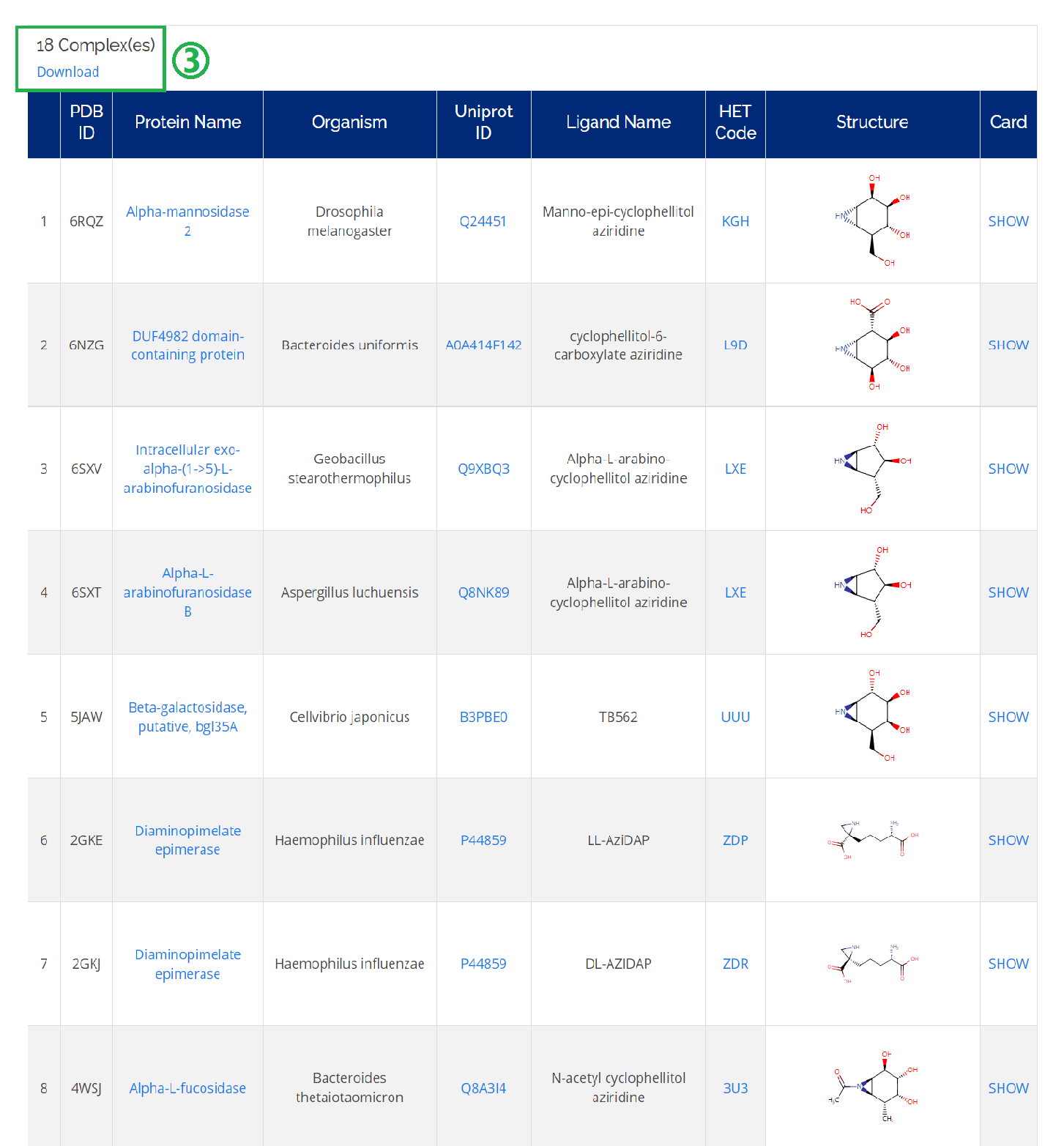
Click on the Complexes icon on the homepage or Home -> Browse -> Complexes.
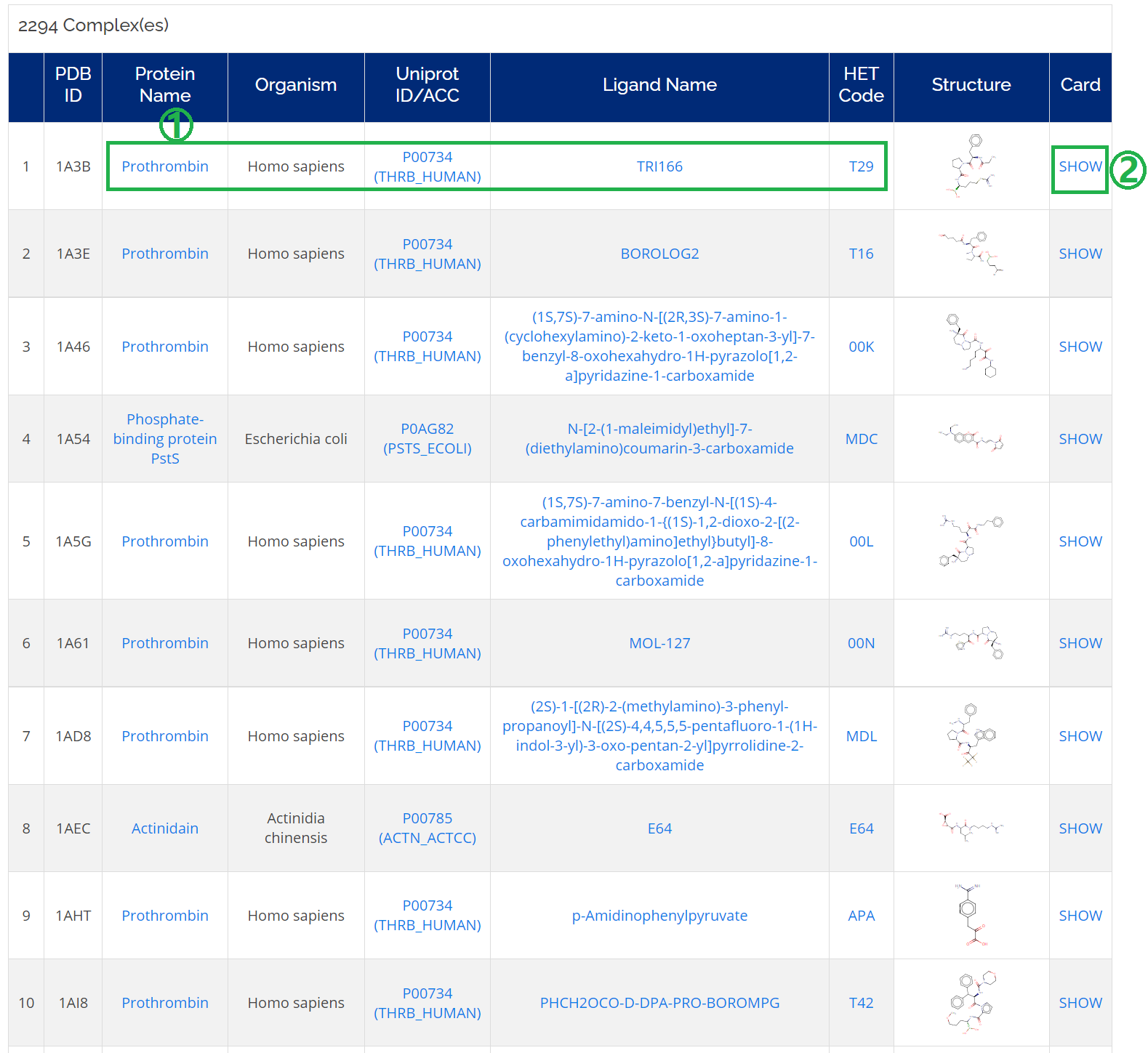
- 1. You could check all the related information (protein name, Uniprot ID, ligand name, and HET code) of a complex of interest;
- 2. Moreover, you could find detailed information on the complex card by clicking SHOW.
.
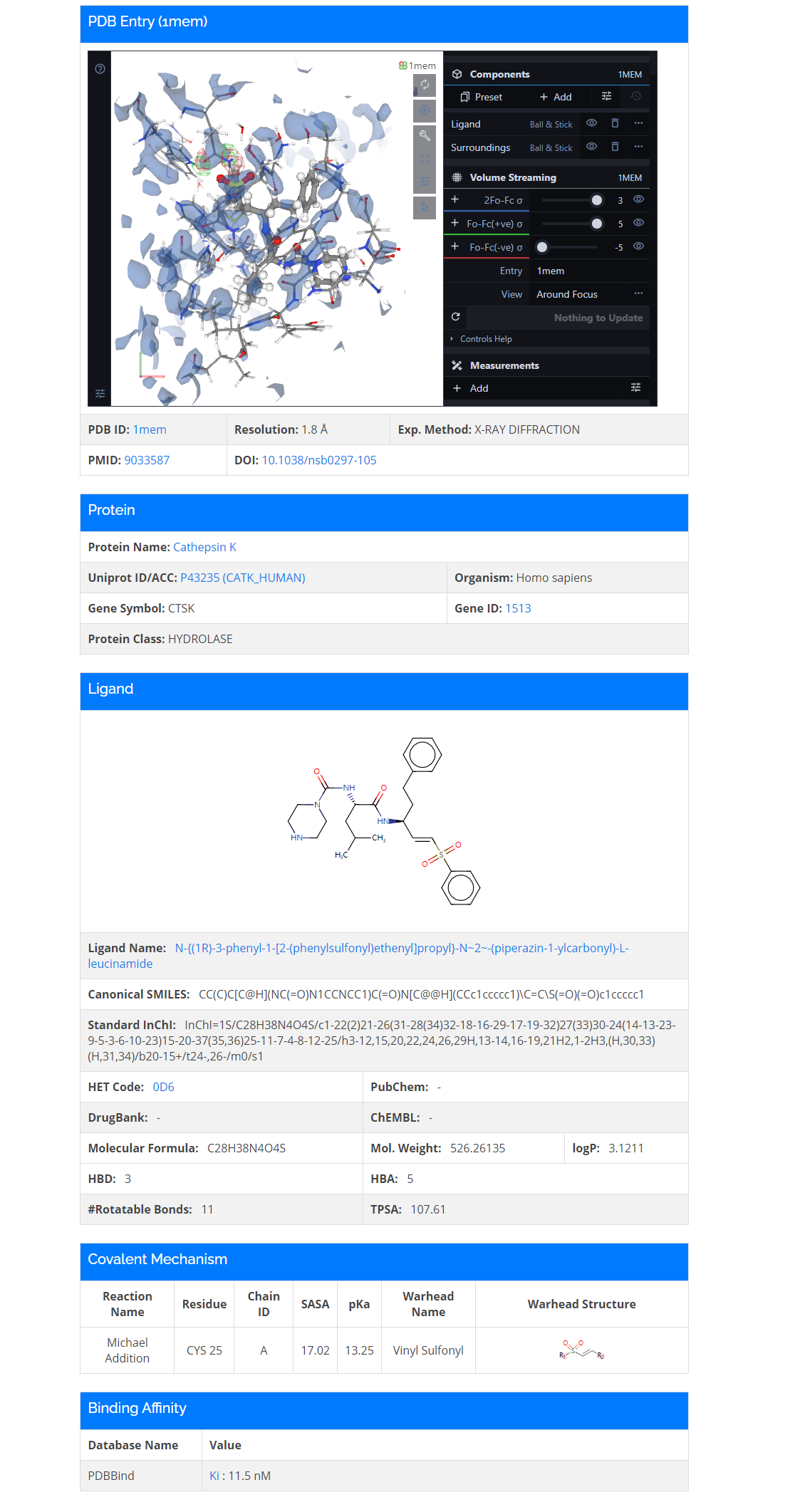
Warheads
There is a Warheads icon on the homepage or you can go Home -> Browse ->Warheads which you could browse all the Warheads SMARTS and Structures that formed covalent bonds with targetable residues.
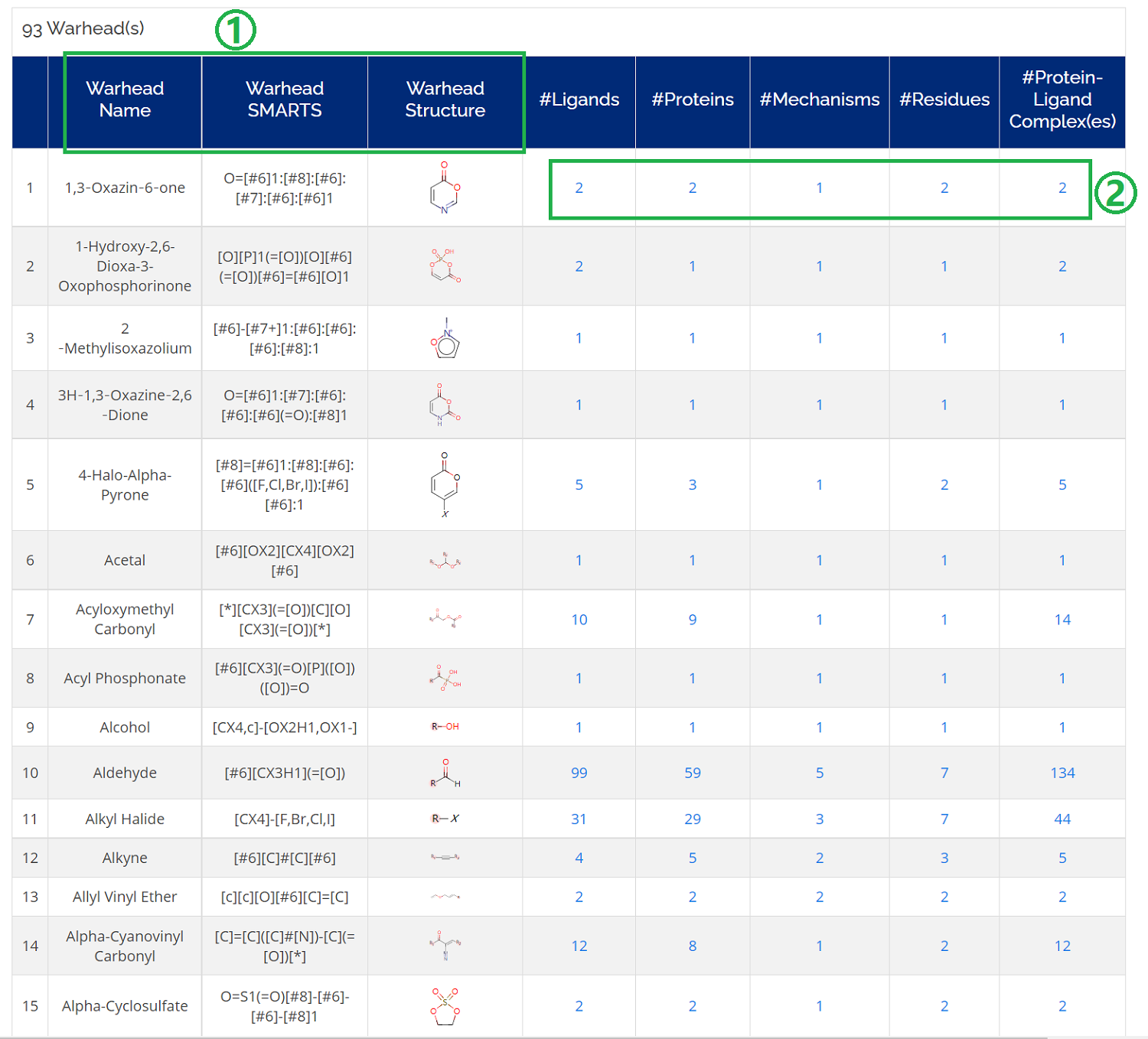
- 1. Warhead name, warhead smarts, and warhead structure are displayed on the warhead page;
- 2. You can check the numbers of ligands, proteins, mechanisms, residues, and complexes related to a specific mechanism;
- 3. More importantly, you could download further information by clicking the number of each term. For example, you could download the ligand information of a residue of interest by clicking a number of #Ligand.
Covalent Mechanisms
Click on the Mechanisms icon on the homepage or Home -> Browse -> Cov. Mechanisms. A table will be displayed for all the 21 different mechanism types.
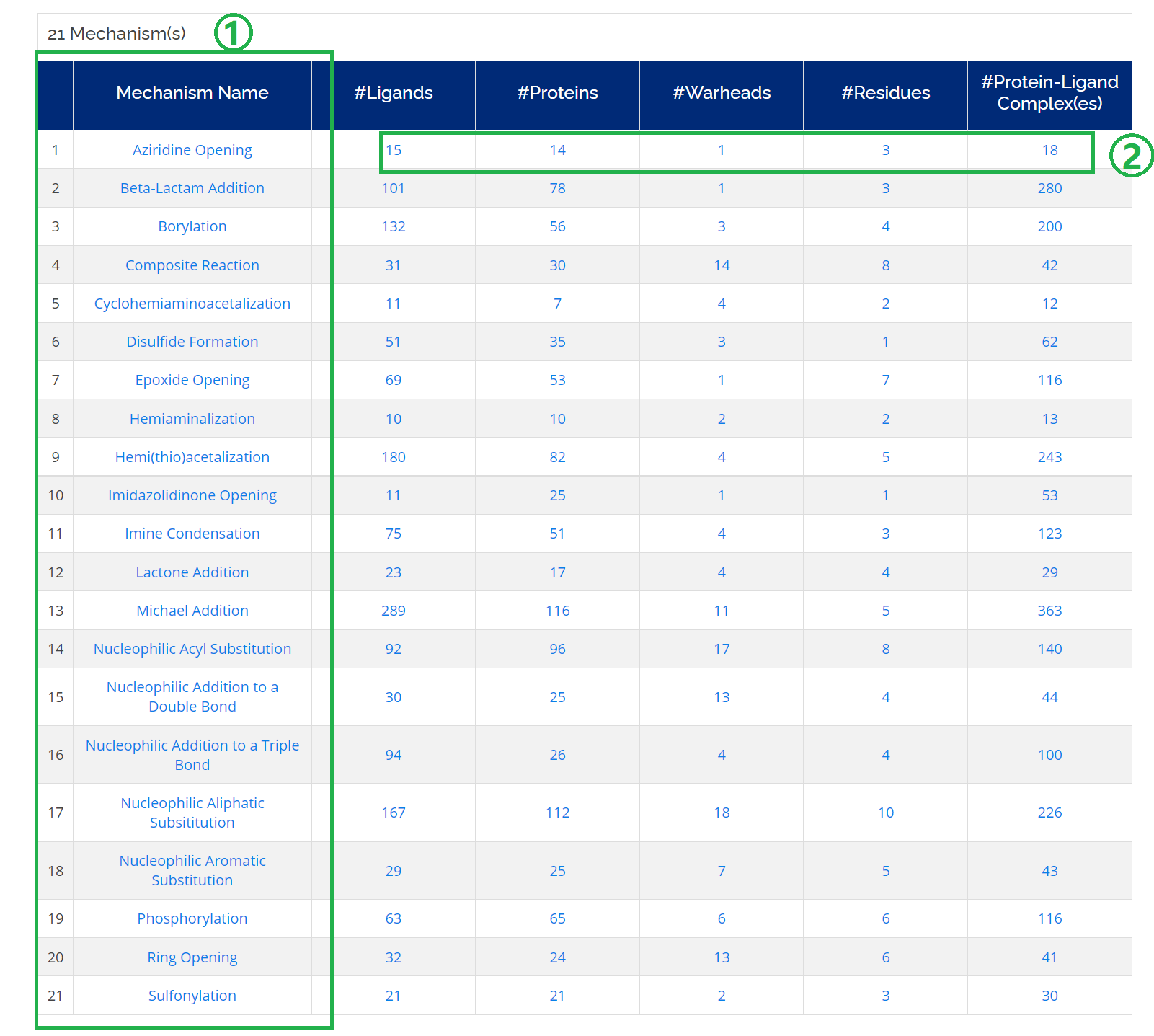
- 1. Detailed information will be opened by clicking one of the Mechanism Name;
- 2. You can check and dowmload detailed information of ligands, proteins, warheads, residues, and complexes related to a specific covalent mechanism.
- 3. For example, you could download the complexes information of a mechanism of interest by clicking a number of #Protein-Ligand Complex(es).
Targetable Residues
Click on the Residues icon on the homepage or Home -> Browse -> Targetable Residues. A table will be displayed for all the 14 different targetable residues.
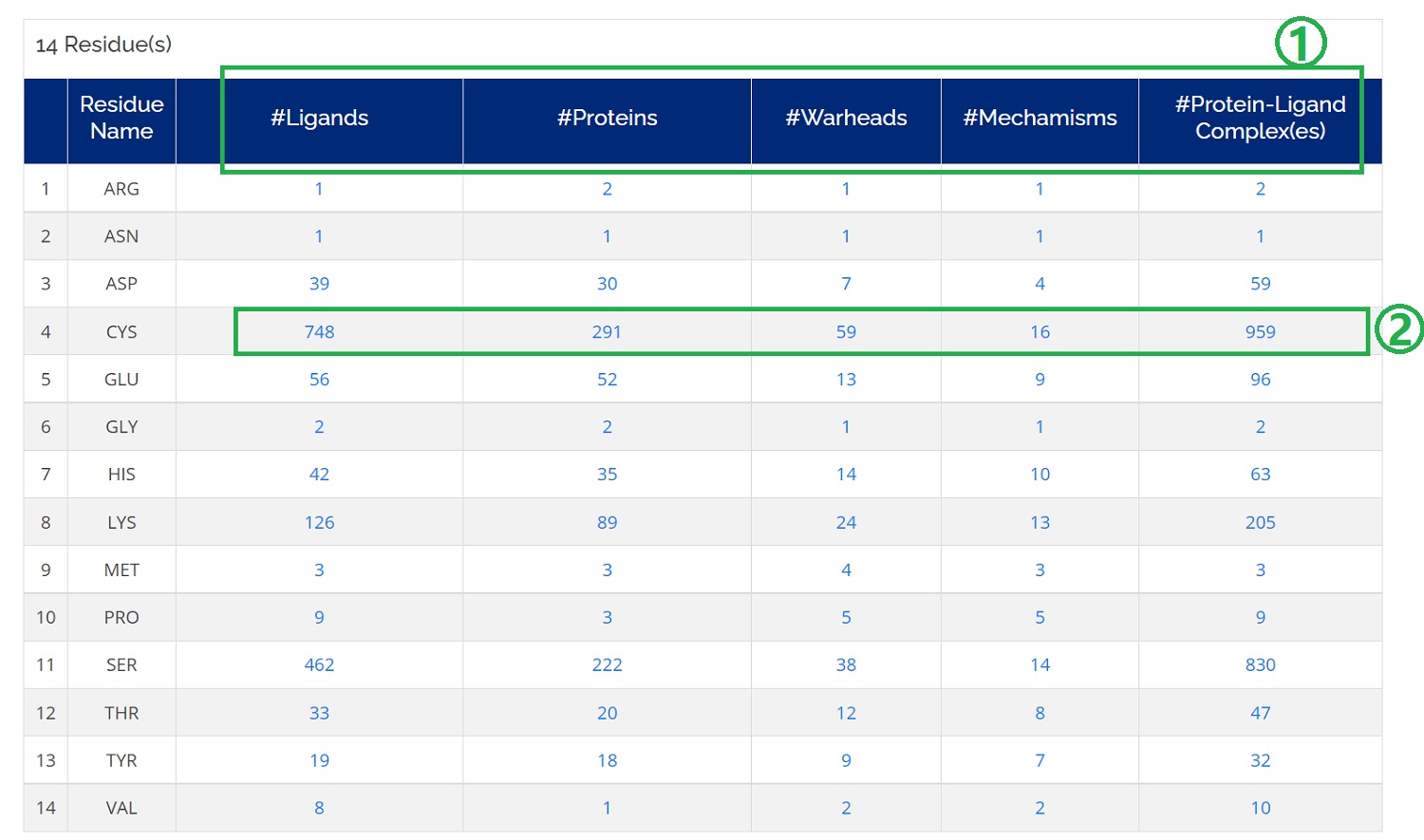
- 1. The targetable residues table contains valueble information, such as ligands, proteins, warheads, mechanisms, and complexes;
- 2. You could click the different number of all the related information for a targetable residue of interest;
- 3. For example, you could download all the complexes by clicking the number of protein-ligand complexes for a specific residue type.
Search CovPDB

Ligand Name/HET Code/Ligand ID
If you know the name of the ligand of interest, you could go Home -> Search ->Name/ID ->Ligand Name/HET Code/Ligand ID, then just input the HET Code, ligand name, or synonym.
Sometimes, the name of a compound is not clearly defined or not known, but the structure of the compound is known. In that case, it is possible to draw the structure by using the ChemDoodle structure editor. It is not necessary to match the exact structure, similar structures will also be identified.
Protein Name/Uniprot ID
If you know the name of the protein of interest, sers can try Home -> Search ->Name/ID ->Protein Name, then just input the protein name or synonym.
Or if you know the UniProt ID or Entry Name of the protein of interest, you can also search through Home -> Search ->Name/ID ->Uniprot ID/ACC.
PDB ID
If you know the PDB ID of the complex of interest, you can try Home -> Search ->Name/ID ->PDB ID, then just input the PDB ID and will be led to a complex card of this PDB entry.
Sequence
Sequence Similarity Searching of CovPDB uses BLAST+ tool to perform fast local alignment searches for a query sequence. For a sequence of interest, navigate to Home -> Search -> Name/ID -> Sequence, just type in a query sequence without header.
The search result page displays an overview of all the retrieved hits with their corresponding scoring. Below the table, a more detailed alignment result is displayed.
Ligand Similarity Search
It is possible to change the search parameters e.g. decreasing the tanimoto coefficient in order to increase the number of retrieved similar compounds. Additionally, you can decide to search for substructures, e.g. you can draw a tricyclic structure to retrieve all tricyclic compounds contained in the CovPDB database.
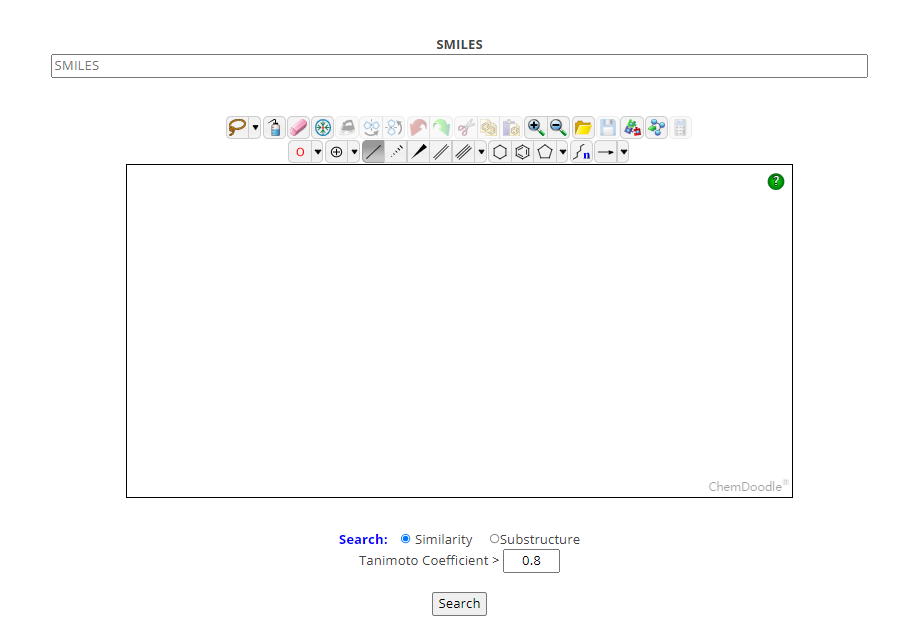
Ligand similarity searches in CovPDB are conducted with RDKit using Morgan fingerprints or substructure searches. Those fingerprints represent structural features of a molecule. Fingerprints are dichotomous [0,1] (bit) arrays. CovPDB uses the tanimoto coefficient for similarity calculations:
tcA,B=NA,B/(NA+NB-NA,B), with
- NA number of bits set "on" in fingerprint of molecule A
- NB number of bits set "on" in fingerprint of molecule B
- NA,B number of bits set "on" in fingerprint of both molecules (A and B).
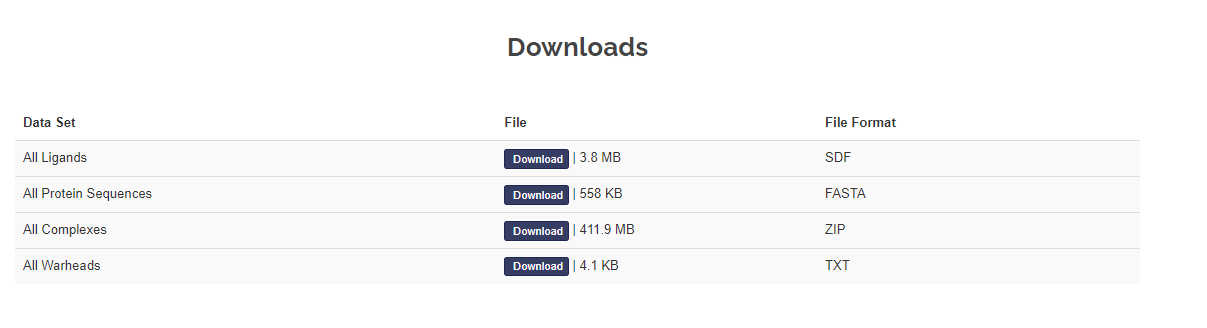
Download
CovPDB freely provides its content as separate files for download, namely, all ligand structures (SDF format), protein sequences (FASTA format)protein-ligand complexes (PDB format), and reactive warheads (TXT format).
Linked Resources
CovPDB cites data from the following public resources:
- ChEMBL
- PubChem
- DrugBank
- Universal Protein Resource (UniProt)
- Protein Data Bank in Europe (PDBe)
- Protein Data Bank (PDB)
- NCBI Gene
- BindingDB
- PDBbind
- Binding MOAD
FAQs
What is the COVPDB ID?
The COVPDB ID is a unique ID that has been assigned to ligands in CovPDB. It can be used to retrieve a Ligand Card page for these entities.
How often is the data updated?
The data of CovPDB will be updated periodically to include more entries upon the availability of newly deposited 3D models in the PDB.
What can CovPDB help with research?
To the best of our knowledge, CovPDB is the first freely accessible web database solely dedicated to high-resolution 3D structures of biologically relevant covalent protein–ligand complexes, mined from the Protein Data Bank. CovPDB describes 2,294 covalent complexes, 1,501 covalent ligands, 733 associated protein targets, 93 reactive warheads, 21 covalent mechanisms, and 14 targetable residues.
The application of quantum chemistry to study the mechanism of action of covalent complexes is a general and versatile approach. We have collected 93 different warheads and high-resolution PDB structures in our database. Therefore, researchers can use the data from CovPDB to apply the QM/MM or QM method to study enzyme reaction and warhead reactivity.
Covalent ligands/inhibitors are considered to be an important part of drug discovery and therapeutics. Thousands of co-crystal structures deposited in CovPDB provide comprehensive information for structure-based covalent ligand design for a protein of interest. Different covalent docking tools are be used to do covalent docking. You could find the high-resolution 3D covalent protein structures from the Download section which can be directly applied to a covalent docking.
Conserved sequence motifs and homology modeling studies could be performed with the protein sequences.
How were the SASA and pKa values of the targetable residues calculated?
SASA and pKa values were calculated with FreeSASA and PROPKA programs, respectively.
FreeSASA is an open source library for calculating solvent accessible surface areas (SASA) of protein, RNA and DNA molecules. We calculated SASA of targetable residues directly from PDB files using default parameters.
PROPKA (version 3) predicts the local pKa values of targetable amino acid residue of protein ionizable groups depending on the desolvation and dielectric response for the protein.
Cite Us
Mingjie Gao, Aurélien F A Moumbock, Ammar Qaseem, Qianqing Xu, Stefan Günther
CovPDB: a high-resolution coverage of the covalent protein–ligand interactome, Nucleic Acids Research, 2021.